Explore DIPY
Command-Line Interfaces
All the algorithms are available using CLI. You can also create your own algorithms.
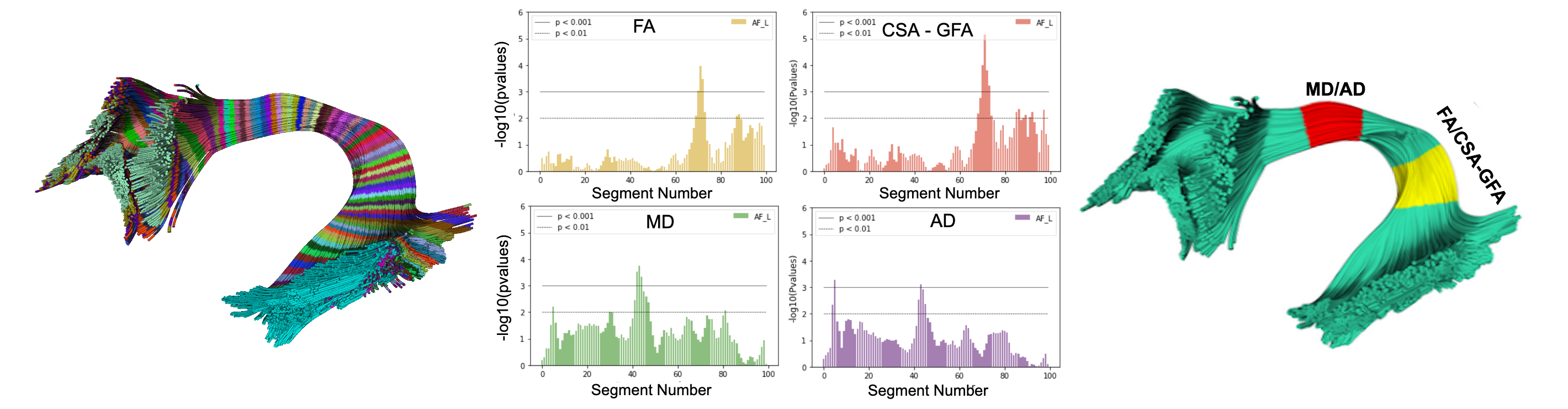
Know moreStatistical Analysis
DIPY allows you to use different methods like BUAN, AFQ, K-Fold cross validation.
Know moreReconstruction
Single shell: DTI, CSA, SFM, SDT, Q-Ball, CSD, and many more.
Multi shell: GQI, DTI, DKI, SHORE, MAPMRI, MSMT-CSD, and many more.
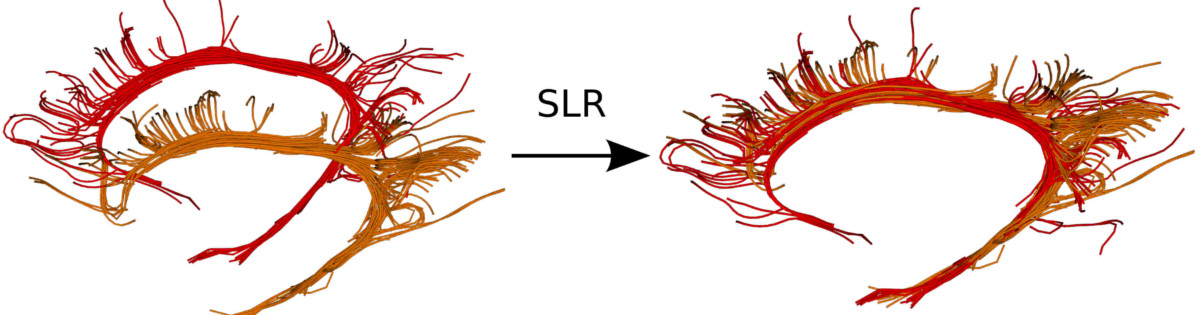
Registration
Affine Registration, Diffeomorphic 2D/3D Registration, Streamlines based Registration, and much more.
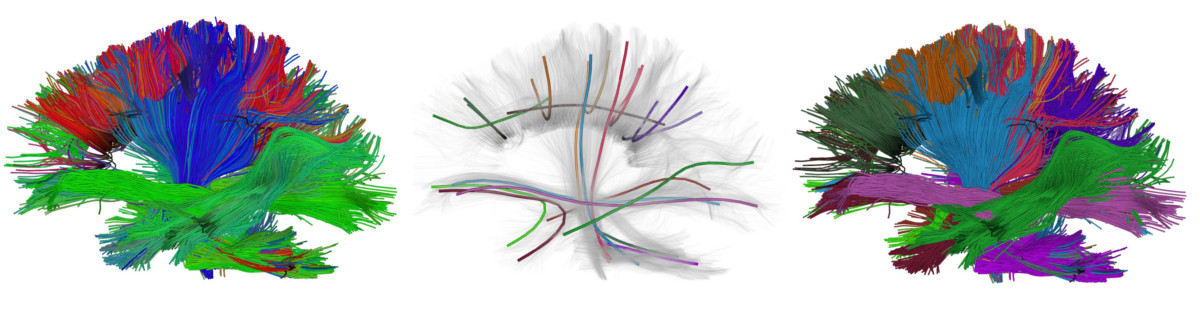
Know moreVisualization
ODFs visualization, interactive tractogram visualization, Advanced UI and Shaders, and much more.
Know moreCite Us!
DIPY, a library for the analysis of diffusion MRI data.
Garyfallidis E, Brett M, Amirbekian B, Rokem A, van der Walt S, Descoteaux M, Nimmo-Smith I and Dipy Contributors (2014).
Frontiers in Neuroinformatics, vol.8, no.8.