Google Summer of Code 2018#
Introduction to DIPY#
DIPY is a free and open source software library for computational neuroanatomy and medical data science. DIPY contains algorithms for diffusion magnetic resonance imaging (dMRI) analysis and tractography but also contains implementations of other computational imaging methods such as denoising and registration that are applicable to the greater medical imaging and image processing communities. Additionally, DIPY is an international project which brings together scientists across labs and countries to share their state-of-the-art code and expertise in the same codebase, accelerating scientific research in medical imaging. DIPY is participating in GSoC this year for the 3rd time under the umbrella of the Python Software Foundation (PSF).
How to become a part of DIPY’s Google Summer of Code 2018#
GSoC is a program that allows students to learn by contributing to an open-source project, while receiving a fellowship from Google, and mentorship from open-source software developers. For details about this year’s GSoC, please refer to this page.
Before considering becoming part of the DIPY GSoC, please read about our expectations.
All participants should have basic knowledge of scientific computing and development in Python. For a comprehensive introduction to these topics, please refer to the book Effective Computation in Physics by Katy Huff and Anthony Scopatz. However, you should be already familiar with data analysis using Python and Numpy before applying.
Be happy to ask questions directly in our Gitter channel https://gitter.im/nipy/dipy
Advice#
Potential candidates should take a look at the guidelines on how to contribute to DIPY. Making a small enhancement/bugfix/documentation fix/etc to DIPY already before applying for the GSoC is a requirement from the PSF; it can help you get some idea how things would work during the GSoC. The fix does not need to be related to your proposal. We have and will continue adding some beginner friendly issues in github.
Projects#
DIPY workflows and Quality Assurance
Description: Create new dipy.workflows and make those executable in different platforms. DIPY has a unique system that allows to create command line interfaces in a systematic and precise way to run across platforms. DIPY uses existing technology such as the default argument parser of Python but enhances the parser using a software engineering process called introspection. Our IntrospectiveParser allows to generate workflows that can be executed both by the command line and using Python scripts. In this work, you will have to:
Take existing tutorials and generate new workflows from them. Test the workflows with new data and generate automated reports.
Help with simplifying installation in the different operating systems.
Difficulty: easy to intermediate
Skills required: Numpy, Python, pyinstaller (or similar), medical imaging.
Mentors: Serge Koudoro and Eleftherios Garyfallidis
Extend Visualization - Focus in UI
Description: In this project you will build scifi-like 3D and 2D user interfaces inspired from Guardians of the Galaxy video. Dipy.viz provides many visualization capabilities. However we were not happy with interactive capabilities found in existing GUIs. For this reason we built our own UI engine. No Qt! Everything is integrated in the VTK scene. See example below that was generated during our 2016 GSoC participation. This is an example of an orbital orbital menu.
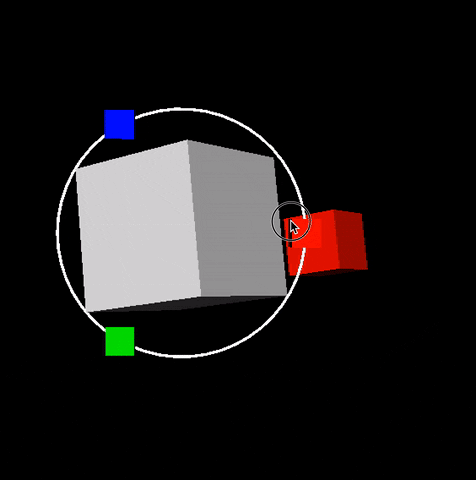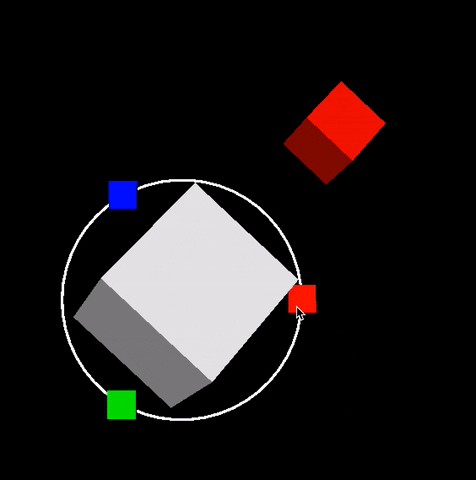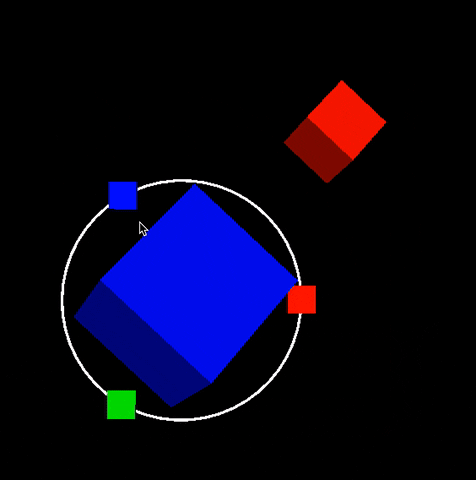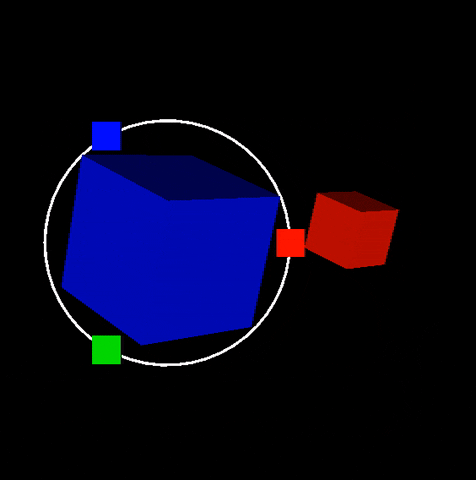
In this project you will extend this work and add more futuristic widgets. The motto of this project is make everything interactive without performance issues. See also figure of Project 5.
Difficulty: intermediate
Skills required: Python, OpenGL and VTK
Mentors: David Reagan and Ranveer Aggarwal and Eleftherios Garyfallidis
Improve performance of nonrigid image registration
Description: We have some really nice code for nonrigid registration that needs to be parallelized. The code is written in Python and Cython. You will need to work primarily on adding multithreading (OpenMP) capabilities in our Symmetric Normalization framework. Start by playing with the following tutorials
Difficulty: intermediate.
Skills required: Familiarity with OpenMP, Cython, Python, Numpy.
Mentor: Serge Koudoro and Eleftherios Garyfallidis
Extend Clustering Framework
Description: QuickBundles and QuickBundlesX are extremely fast algorithms that can be used in a series of fields and datasets. We initially used this algorithm to cluster streamlines. Your job will be to extend our existing framework to new datasets. For, example implement new metrics that allow to cluster surfaces, images, text or other. Also, you will have to work on a research component of the algorithm that is related to reducing the number of clusters in dense datasets.
Difficulty: intermediate
Skills required: Python/Cython, machine learning. Especially, unsupervised learning. Knowledge of scikit- learn is an advantage.
Mentor: Eleftherios Garyfallidis and Serge Koudoro
Extend Visualization - Focus in GLSL
Description: Our new visualization engine supports GLSL shading language. Join our effort to built stunning visualizations of brain images and other scientific datasets. You will have to program vertex and fragment shaders to generate different effects on VTK polydata. For examples, see code here. Here is an example without shaders
You will have to update the code to enable shading when needed and if supported by the current computing system. Please also check tutorials starting with viz here
Difficulty: high
Skills required: GLSL, Python, OpenGL and VTK
Mentors: David Reagan, Ranveer Aggarwal and Eleftherios Garyfallidis
Implement new models for microstructure imaging
Description: This is a model fitting project. You will be required to extend our new microstructure framework. You will be able to implement models such as Multi Tensor, NODDI, Axcaliber, CHARMED, Ball & Sticks, Ball & Rackets all with three crossings and also all the combinations of Zeppelin, Cylinder, Dot and Ball compartments. See MRI example below. How would you model these tiny structures?

Difficulty: high
Skills required: MSc or PhD level, mathematical optimization, Python, Numpy, Cython (bonus)
Mentors: Maryam Afzali, Mauro Zucchelli and Eleftherios Garyfallidis
Extend and QA tracking framework
Description: Tractography is one of the great challenges in medical imaging. In DIPY we have implemented different tracking algorithms including deterministic, probabilistic and particle filtering algorithms. You will have to extend dipy.tracking with machine learning based algorithms. Also you will need to test your algorithm with different datasets of different resolutions.
Difficulty: high
Skills required: Python/Cython, knowledge of tractography. Available only for MSc or PhD students.
Mentor: Eleftherios Garyfallidis, Serge Koudoro, Gabriel Girard and Ariel Rokem.