Time to start mapping brain connections and looking to brain properties in vivo#
Hi all,
Tomorrow we are starting the coding period :), so it is time for some details about my project and tell you what was done in the community bonding period.
How can we study brain connections and brain’s tissue properties in vivo? - A simple introduction for non experts
Trajectory of neuronal connections (tractography) and quantification of tissue properties in the living human brain can be obtain from measures of water diffusion using MRI scans. To give you an example how this is done, I will first start by describing one of the simplest technique - the diffusion tensor imaging (DTI).
By combining the information of several diffusion weighted images, DTI models the water diffusion for each image element using a tensor which can be represented by an ellipsoid (see Figure below).
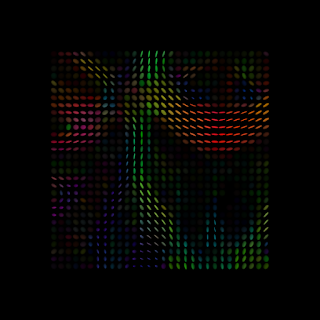
Figure 1. Diffusion tensors computed from all voxels of a real brain image. This image was produced using Dipy as described in Dipy’s website.#
From figure 1 we can see that diffusion is larger is some directions. In fact the direction of larger diffusion can be related to the direction of brain’s white matter fibers. The axon myelin sheaths restricts the water diffusion and thus diffusion is smaller on the directions perpendicular to fibers. On the other hand, the diffusion parallel to fibers is less restricted and therefore matching the direction of fibers.
Based on this, 3D virtual reconstruction of brain connection can be obtain using specific tracking algorithms - a procedure which is named fiber tracking. An example of this 3D maps obtain from a real brain dataset is shown below.
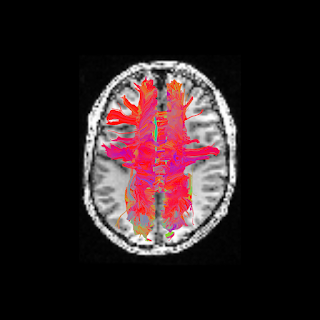
Figure 2. Example of corpus callosum fibers. These fibers connect the left and right fiber hemispheres. This image was produced using Dipy as described in Dipy’s website.#
Nowadays, DTI is still one of the diffusion weighted techniques most used in both clinical applications and in many research studies, however it is not always accurate. DTI cannot account properly for the crossing of different populations of white-matter fiber connections. Moreover, it ignores the non-Gaussian properties of diffusion in biological tissues which can be used to derive interesting and important measures of tissue properties.
Project proposal
In this project, I will be implementing an alternative the diffusion-weighted technique named the diffusion kurtosis imaging (DKI) in an open source software project, the Diffusion Imaging in Python (Dipy). DKI overcomes the two major limitations of DTI: It quantifies the non-Gaussian properties of water diffusion in biological tissues by modelling the kurtosis tensor (KT) which can be used to derive important tissue measures as the density of axonal fibers. Relative to the diffusion tensor, KT is also shown to offer a better characterization of the spatial arrangement of tissue microstructure and can be used as a basis for more robust tractography. Particularly, DKI based tractography is sensitive to resolve crossing fibers.
What is done so far
As an update of what I posted previously (see Post #2), I finished the work on DKI’s simulations - procedures that will be useful for testing the codes that I will be implementing during this summer. In particular, as my mentor suggested, I added some automatic debugging scripts using Nose python testing. These scripts are now insuring that the kurtosis tensor is symmetry (as expected) and that simulations are able to currently produce the diffusion tensor and kurtosis tensor in both cases of well aligned and crossing fibers.
Many thanks to my mentor for teaching me how to work with nose python testing. In particular, the useful tip running the nose tests and knowing which lines the testing scripts are covering by using the following command:
1 nosetests -v dipy/sims/tests/test_voxel.py --with-coverage --cover-package=dipy
Next steps
After merging the DKI simulations to Dipy’s master branch, I will start working on the DKI reconstruction modules, based on some preliminary preparation work previously submitted by other dipy contributors. At the end of the week, I intend to finish the first part of the DKI reconstruction modules - the KT estimation from diffusion-weighted signals. For this I will implement the standard ordinary linear least-squares (OLS) solution of DKI.