Mid-Term Summary#
We are now at the middle of the GSoC 2015 coding period, so it is time to summarize the progress done so far and update the plan for the work of the second half part of the program.
Progress summary#
Overall a lot was achieved! As planned on my project proposal, during the first half of the coding period, I finalized the implementation of the first version of the diffusion kurtosis imaging (DKI) reconstruction module. Moreover, some exciting extra steps were done!
Accomplishing the first steps of the project proposal#
The first accomplished achievement was merging the work done on the community bonding period to the main master Dipy repository. This work consisted on some DKI simulation modules that can be used to study the expected ground truth kurtosis values of white matter brain fibers. In this project, these simulations were useful to test the real brain DKI processing module. The documentation of this work can be already found in Dipy’s website.
The second achievement was finalizing the procedures to fit the DKI model on real brain data. This was done from inheritance of a module class already implemented in Dipy, which contains the implementation of the simpler diffusion tensor model (for more details on this you can see my previous post). Completion of the DKI fitting procedure was followed by implementation of functions to compute the ordinary linear least square fit solution of the DKI model. By establishing the inheritance between the DKI and diffusion tensor modules, duplication of code was avoided and the standard diffusion tensor measures were automatically incorporated. The figure below shows an example of these standard measures obtained from the new DKI module after the implementation of the relevant fitting functions.
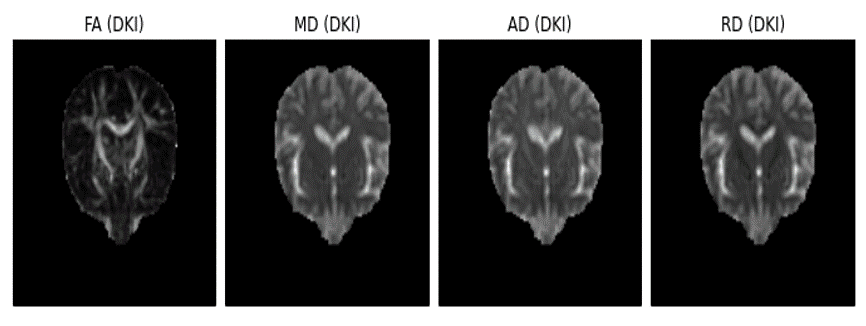
Figure 1 - Real brain standard diffusion tensor measures obtained from the DKI module, which included the diffusion fractional anisotropy (FA), the mean diffusivity (MK), the axial diffusivity (AD) and the radial diffusivity (RD). The raw brain dataset used for the images reconstruction was kindly provided by Maurizio Marrale (University of Palermo).#
Finally, from the DKI developed fitting functions, standard measures of kurtosis were implemented. These were based on the analytical solutions proposed by Tabesh and colleagues which required, for instance, the implementation of sub-functions to rotate 4D matrices and to compute Carlson’s incomplete elliptical integrals. Having implemented the analytical solution of standard kurtosis measure functions, I accomplished all the work proposed for the first half of the GSoC. Below I am showing the first real brain images reconstructed kurtosis from the new implemented modules.
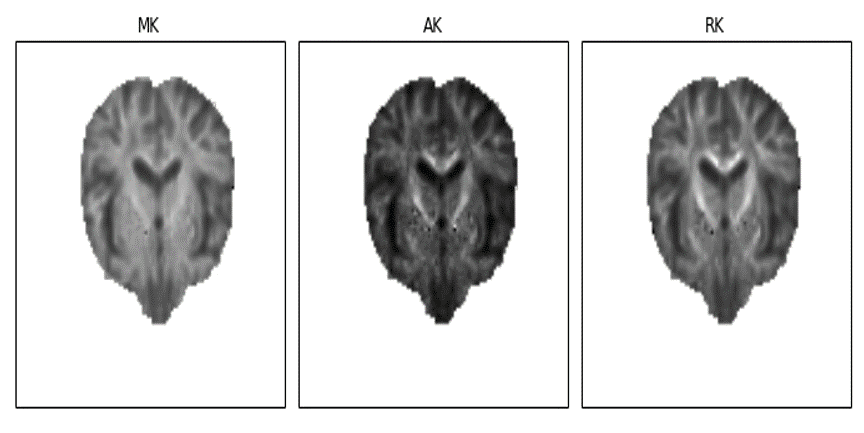
Figure 2 - Real brain standard kurtosis tensor measures obtained from the DKI module, which included the mean kurtosis (MK), the axial kurtosis (AK), and radial kurtosis (RK). The raw brain dataset used for the images reconstruction was kindly provided by Maurizio Marrale (University of Palermo).#
Extra steps accomplished#
Some extra steps were also accomplished during the first half of the GSoC program. In particular, from the feedback that I obtained at the International Society for Magnetic Resonance in Medicine (ISMRM) conference (see my fourth post), I decided to implement an additional DKI fitting solution - the weighted linear least square DKI fit solution. This fit is considered to be one of the most robust fitting approaches in recent DKI literature (for more details see my previous post). Therefore, having this method implemented, I am insuring that the new Dipy’s DKI modules are implemented according to the most advanced DKI state of art.
To show how productive was the ISMRM conference for the project, I am sharing you a photo that I took at the conference with one of the head developers of Dipy - Eleftherios Garyfallidis.
Figure 3 - Photo taken at the ISMRM conference - I am wearing the Dipy’s T-shirt at the right side of the photo and in the left side you can see the head Dipy’s developer Eleftherios Garyfallidis.#
Next steps#
After discussing with my mentor, we agreed that we should dedicate more time on the first part of the project proposal, i.e. improving the DKI reconstruction module. Due to the huge extent of code and the math complexity of this module, I will dedicate a couple of weeks more in improving the module’s performance, quality of code testing and documentation. In this way, we decided to postpone the two last milestones initially planned for the second half term of the GSoC to the last three weeks of the GSoC coding period.
The next steps of the updated project plan are as described in the following points:
Merge the pull requests that contain the new DKI modules with the master’s Dipy repository. To facilitate the revision of the implemented functions by the mentoring organization, I will split my initial pull request into smaller pull requests.
At the same time that the previous developed code is reviewed, I will implement new features on the functions for estimating kurtosis parameters to reduce processing time. For instance, I will implement some optional variables that allow each method to receive a Boolean mask to point the image voxels to be processed. This will save the time wasted on processing unnecessary voxels as from the background.
I will also implement simpler numerical methods for a faster estimation of the standard DKI measures. These numerical methods are expected to be less accurate that the analytical solutions already implemented, however they provide alternatives less computationally demanding. Moreover, they will provide a simpler mathematical framework which will be used to further validate the analytical solutions.
Further improvements of the weighed linear least square solution will be performed. In particular, the weights’ estimations used on the fit will be improved by an iterative algorithm as described on recent DKI literature.
Finally, the procedures to estimate from DKI concrete biophysical measures and white matter fiber directions will be implemented as I described on the initial project proposal.