Artifacts in Dipy’s sample data Sherbrooke’s 3 shells#
Hi all,
Just to report an issue that I am currently trying to figure out!
As I showed on my previous post my first diffusion kurtosis reconstructions are looking very good. However when I try to process the Dipy’s multi-shell dataset sample Sherbrooke 3 shells, kurtosis measures seem to be widely corrupted by implausible high negative values see figure below:
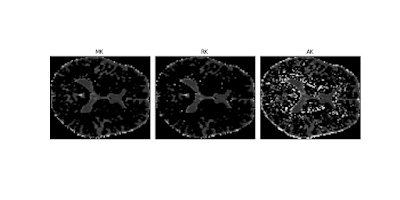
Fig.1 - Diffusion kurtosis standard measures obtain from the Sherbrooke 3 shells Dipy’s sample dataset.#
Mid-Term Summary
Perpendicular directions samples relative to a given vector