Progress Report on Diffusion Kurtosis Imaging (DKI) Implementation#
We are almost getting to the end of the GSoC coding period 😭.
The good news is that progress is still going at full speed!!! I finalized the work on the standard kurtosis statistics estimation, and great progress was done on the white matter fiber direction estimates from diffusion kurtosis imaging (DKI). Details can be found below!
Implementation of DKI statistics is now complete!!!#
As I planned in my previous post, in the last couple of weeks, I created a sample usage script for the DKI statistic estimation module using data acquired with similar parameters to the Human Connectome Project. Figures, for both diffusion and kurtosis standard statistics are looking very good (see below) and these are great news. These results show that my implemented module can be used on the analysis of one of the largest worldwide projects which aims to map the human brain connections.
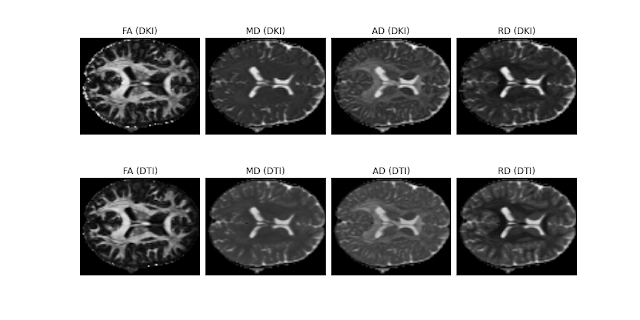
Figure 1. Real brain parameter maps of the diffusion fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD) obtained from a HCP-like dataset using the DKI modules (upper panels) and the DTI module (lower panels). Despite DKI involves the estimation of a larger number of parameters, the quality of the diffusion standard measures of the HCP-like dataset from DKI seem to be comparable with the standard diffusion measures from DTI. This dataset was kindly supplied by Valabregue Romain, CENIR, ICM, Paris.#
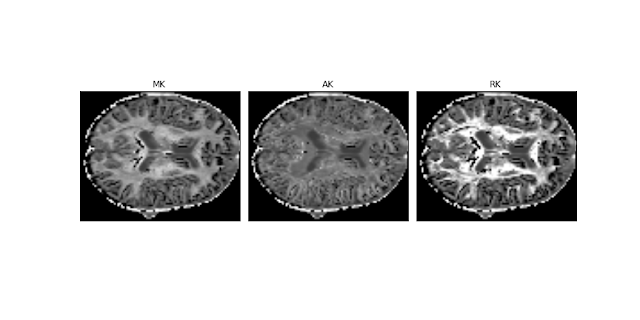
Figure 2 - Real brain parameter maps of the mean kurtosis (MK), axial kurtosis (AK), and radial kurtosis (RK) obtained from a HCP-like dataset using the DKI module. These are the maps specific to DKI. The dataset for these reconstructions was kindly supplied by Valabregue Romain, CENIR, ICM, Paris.#
I also dramatically improved the speed performance of the kurtosis statistics estimation modules! In my previous post, I mentioned that I had optimized the codes in the way that all three standard kurtosis statistics are processed within 30 min. Now all three standard kurtosis statistics can be computed within 1 min. Reprocessing the kurtosis measures shown in Figure 1 of my post #6 is now taking:
Mean kurtosis - 32 sec (before 14 mins)
Radial kurtosis - 12 sec (before 7 mins)
Axial kurtosis - 42 sec (before 1 min)
Advances on the DKI based fiber direction estimates#
Based on the DKI-ODF described in my previous post, a procedure to extract the fiber direction estimates was implemented. This was done using the quasi-Newton algorithms available on Scipy’s optimization module. For an example of the fiber direction estimates using the implemented procedure, we show above the estimates obtained from real brain voxels of the corpus callosum:
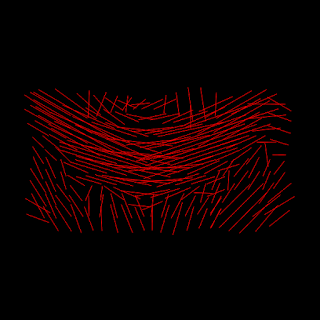
Figure 3 - Sagittal view of the direction estimates of horizontal corpus callosum fibers obtained from the DKI-ODF.#
Last steps for the Google Summer of Code 2015#
The work on the DKI based fiber direction estimates will be finalized by making the fiber estimates compatible with the tractography methods already implemented in Dipy. In this way, I will be able to reproduce the first DKI based tractography in HCP-like data.
With the procedures to estimate the standard kurtosis statistics and DKI based fiber estimates, I will finish the work proposed in my proposal by implementing some novel DKI measures which can be related to concrete biophysical parameters.