Further improvements on the diffusion standard statistics#
As I mentioned in my last post, I used the implemented modules to process data acquired with similar parameters to one of the largest worldwide projects, the Human Connectome project. Considering that I was fitting the diffusion kurtosis model with practically no pre-processing steps, which are normally required on diffusion kurtosis imaging, kurtosis reconstructions were looking very good (see Figure 2 of my last post).
Despite this, some image artifacts were present, likely being a consequence of gibbs artifacts and MRI noise. In particular, some low intensity voxels were present in regions where we expect that MK and RK is high. To correct these artifacts, I decided to add a pre-processing step that denoises diffusion-weighted data (to see the coding details of this, see directly on my pull request).
Before fitting DKI on the denoised data, these are the amazing kurtosis maps that I obtained:
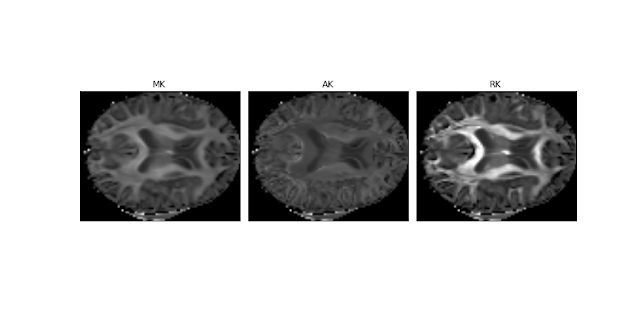
Figure 1 - Real brain parameter maps of the mean kurtosis (MK), axial kurtosis (AK), and radial kurtosis (RK) obtained from a HCP-like dataset using the DKI module. These are the maps specific to DKI. The dataset for these reconstructions was kindly supplied by Valabregue Romain, CENIR, ICM, Paris.#
You can also see the standard diffusion measures obtained from my implemented DKI module and compared to the DTI module previously implemented:
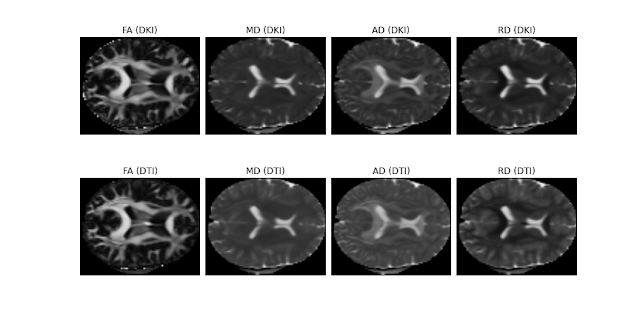
Figure 2. Real brain parameter maps of the diffusion fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD) obtained from a HCP-like dataset using the DKI modules (upper panels) and the DTI module (lower panels). Despite DKI involves the estimation of a larger number of parameters, the quality of the diffusion standard measures of the HCP-like dataset from DKI seem to be comparable with the standard diffusion measures from DTI. This dataset was kindly supplied by Valabregue Romain, CENIR, ICM, Paris.#