Final Project Report#
Hi all!
The GSoC coding period is now over!
Having participated in the GSoC was an amazing experience. In general, all objectives of my project were accomplished. Now, the scientific and wider imaging processing community can have access to the first open source DKI processing modules. As the results of this project showed (see for example my post #10 and post #11), these modules can be used to analyse data of large worldwide collaborative projects such as the Human Connectome Project (HCP). Moreover, I had a great time working with members of my mentoring organization - I learned a lot from them and I will definitely continue contributing to Dipy in the following years.
Below you can find my final project report.
Project summary#
In sum, this project was organized in 4 main phases:
Finishing the work done on functions to simulate signal from the DKI model
Implementing methods for estimating the diffusion kurtosis tensor and derived measures
Adding a procedure to estimate biophysical parameters from DKI
Developing techniques to estimate fiber directions from real DKI data
The details of the work done on each phase is described below:
DKI based simulations#
In this part of the project, I implemented the DKI simulates that were important to test the performance of all functions created in the other steps of the project. Part of this work was done before the GSoC coding period and its finalization was reported in the mid-term summary. Just to highlight the relevance of these simulations, during the GSoC coding period, 19 nose tests functions were created in which 13 were based on DKI based simulates. Moreover, DKI simulations were also useful for selecting, optimizing and debugging DKI methods (see for example post #9 and post #13).
DKI reconstruction modules#
As I proposed in my initial project plan, having a final version of the DKI fitting modules and estimation of diffusion kurtosis statistics was the main goal to achieve for the mid-term evaluation. Since these modules provide the base for the work of the other parts of the project, I decided to dedicate some more time of the second half part of the GSoC coding period to improve the diffusion kurtosis statistics functions. These improvements are summarized in the following points:
The analytical solutions of the mean and radial kurtosis were validated using two numerical methods (post #9).
The performance of the functions were improved so that all standard kurtosis statistics can be computed within 1 min (post #10)
I also explored some Dipy’s pre-processing steps that dramatically improved the quality of the DKI reconstructions (post #11 and post #12).
I added some nosetests to ensure that all code lines of DKI reconstruction modules were covered by nosetest units. From this, I detected some problems with singularities on the function computing the mean kurtosis, which were solved as reported in post #13).
The sample usage script of these modules was adapted according to a new DKI dataset which was required with similar parameters to the HCP.
Below we show the kurtosis statistics images obtained from the HCP-like data using the DKI reconstruction modules before (upper panels of Figure 1) and after (lower panels of Figure 1) the improvements done on the second half part of the GSoC term.
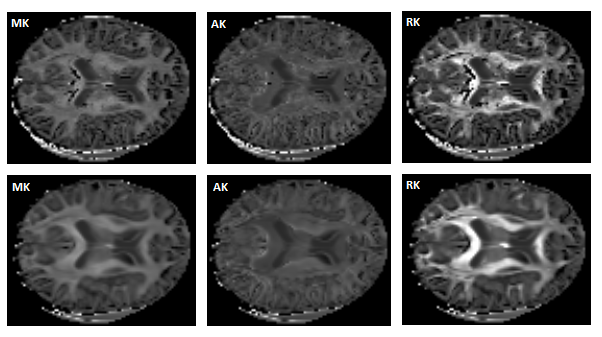
Figure 1 - Diffusion Kurtosis statistics of the HCP-like data obtained from the implemented DKI reconstructions before (upper panels) and after (lower panels) the optimization done on the second half part of the GSoC coding period. Optimized functions seem to correct the artifacts present on the white matter regions as from the splenium of the corpus callosum.#
The final version of the DKI modules can be found in the following pull request.
DKI based biological measures#
Given the extra work done on the previous step, the implementation of the DKI biological measures was rescheduled to the last couple of weeks of the GSoC period. These measures were obtained from the DKI based model proposed by Fieremans et al., (2011), which allows the estimation of concrete biophysical parameters from brain regions of well aligned fibers. Until the end of the coding period, great advances were done on this module. For example, Figure 2 shows the estimated values of axonal water fraction (the proportion of water presented inside the fibers) for voxels containing well-aligned fibers of the splenium and genu of the corpus callosum obtained from the current version of this DKI biophysical model.
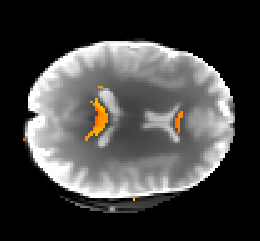
Figure 2 - Axonal water fraction values of the splenium and genu of the corpus callosum (red-yellow colormap values) plotted over the first b-value=0 of the HCP-like diffusion-weighted dataset.#
Unfortunately, since the final version of these functions depends on the other pull requests that are currently being revised, the work done on the implementation of the biophysical models was not finalized, and thus it will not be submitted as part of the GSoC code sample. However, I intend to finalize these codes soon after the GSoC. If you are interested in looking at the final version of the biophysical metric estimations, keep tuned to the updates done at the DKI reconstructions pull request.
DKI based fiber direction estimation methods#
As planned in the project proposal, in the second half part of the GSoC coding period, I developed a procedure to predict the fiber direction estimates from DKI. This was done by first estimating an orientation distribution function (ODF) which gives the probability that a fiber direction is aligned to a specific spatial direction (post #9). From the ODF, fiber directions can be estimated by finding the maxima values of the ODF (post #10). In the last couple of weeks, I accomplished a final version of this procedure by writing its sample usage script, where real brain data ODF and fiber directions are estimated. Visualizations of these estimates are shown in Figure 3 and 4.
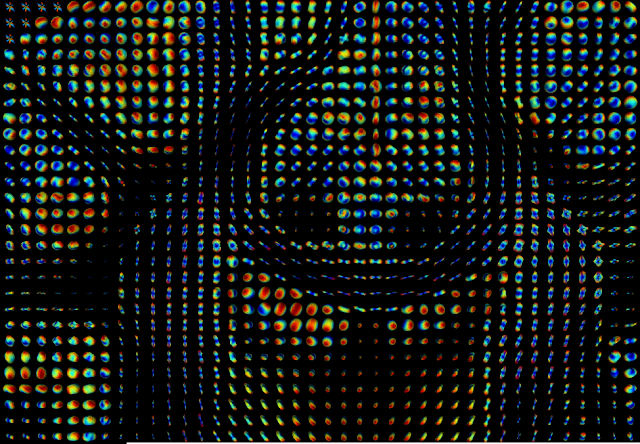
Figure 3 - DKI based orientation distribution function (ODF) computed for voxels of portion of the HCP-like data.#
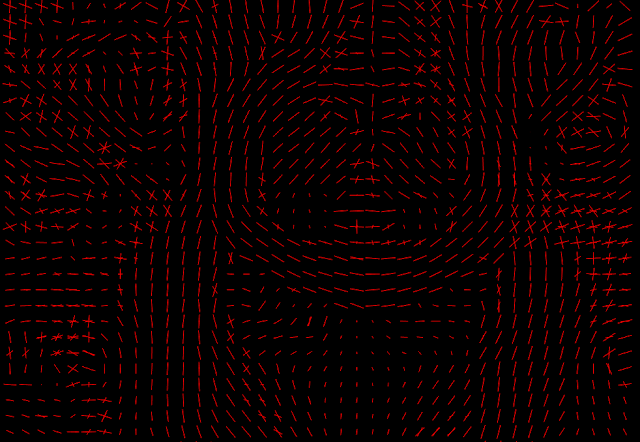
Figure 4 - Fiber directions computed by detecting the directions of maxima ODF. The multiple direction estimates from some voxels show that DKI is able to resolve crossing fibers.#
The final version of the modules containing the function to estimate fiber directions from DKI can be found in the following pull request.
Skills gained on GSoC#
With the supervision of the members of my mentoring organization, I dramatically improved my programming skills.
I learned all required steps to work on collaborative projects such as Dipy. Particularly, I learned how to share, update and comment my work using Github’s development framework.
I learned how to use ipython notebook to create sample script examples, and using ipython profiling techniques to check and improve function performance.
Now I know how to use testing units, such as the nosetest units, which allows me to automatically check bugs on the functions that I am implementing.
I also learned how to improve functions using cython.
Finally, I got familiarized with Dipy’s structure and how to use their functions. This is useful knowledge for my personal future research.